We are grateful to the Super Sam Foundation for supporting Noah as the first Super Sam Fellow. Learn about Sam here. Noah will present the logistics of his work online at this blog – please bookmark it and visit weekly! Here’s Noah:  see also the video about Noah’s work at https://www.facebook.com/SuperSamFoundation/videos/1913393832243243/
see also the video about Noah’s work at https://www.facebook.com/SuperSamFoundation/videos/1913393832243243/
update 6/06/2018:
By popular demand, we are switching to less-frequent, more content-rich monthly posts. These will appear as new blog posts on our website. For exclusive! week to week updates of late-breaking results, email molly@cc-tdi.org to be subscribed. Thank you for supporting this rhabdomyosarcoma research!
——————————————
08/08/2017: Hi everyone! First edition of the cancer math blog. First, a huge thank you to Cassie Santhuff everyone at the Super Sam Foundation, and everyone who supports the amazing work they do to push the world of children’s cancer treatment forward. Without the support of the Super Sam foundation, we wouldn’t be able to do what we do, and I cannot ever thank all of you enough. 
Now, to the science. I’m currently working with a stellar team of interns from Oregon State University to develop a pipeline rapid, high-throughput analysis of sequencing data from pediatric cancers. It’s a privilege to mentor them, and their knowledge expands daily. Excellent taste in coffee, too. This pipeline will reduce the time commitment from a knowledgeable bioinformaticist required to generate standard analysis data, i.e. 40 hours of work per week of active data management will be reduced to a few hours. And that means my work on cancer math continues, as I’m working every day to develop the next iteration of algorithms. I’m excited to engage my colleagues with some of the new capabilities of cancer math, including a standardized approach to disease-wide cancer math modeling, which we will be using as soon as it’s ready.
More to come on cancer math, and this approach to picking the best treatment for an individual, or a group of individuals in need of a relapsed therapy recommendation! 08/30/2017: It’s been a busy couple of weeks. We at cc-TDI had the true privilege of hosting families touched by pediatric cancer and diseases at the 2017 Pediatric Cancer Nanocourse. We welcomed people affected by myriad different diseases; Alveolar Rhabdomyosarcoma, Epithelioid Sarcoma, Bloom’s Syndrome, and Embryonal Tumours with Multilayered Rosettes (ETMR) to name a few. This event occurs every year and the disease focus changes, but the emotional impact is always profound… this chance to connect with parents and families always hits hard, and always strengthens our resolve as individual scientists and as a lab. It’s sometimes hard to believe that I can be a part of something bigger than myself, but spending time with families reminds me that my work in cancer math, and everyone’s work at cc-TDI, offers the promise of new treatments for children’s cancer.  On a lighter note, I’m also happy to report that our SciClone Liquid Handling Robot (a recent major equipment acquisition for cc-TDI) is up and running thanks to the efforts of my parent-turned-scientist/mentee Andy. Here’s us giving a demonstration of this powerful new research tool during the nanocourse to build drug screens on-the-fly in a matter of seconds, a task that previously took hours! I’ll probably brag about Andy a few more times, because mentoring him and working with him is one of my favorite parts of my life at cc-TDI.
On a lighter note, I’m also happy to report that our SciClone Liquid Handling Robot (a recent major equipment acquisition for cc-TDI) is up and running thanks to the efforts of my parent-turned-scientist/mentee Andy. Here’s us giving a demonstration of this powerful new research tool during the nanocourse to build drug screens on-the-fly in a matter of seconds, a task that previously took hours! I’ll probably brag about Andy a few more times, because mentoring him and working with him is one of my favorite parts of my life at cc-TDI. 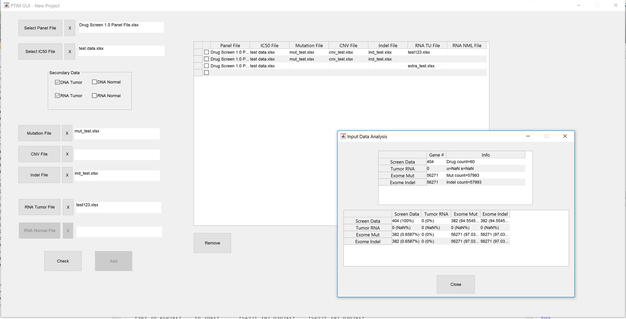 09/14/2017: Stress-testing project building code for the Cancer Math GUI. One of the key aspects of developing software is to make it accessible to everyone. Developing this front-end GUI to enable the whole lab to access Cancer Math algorithms quickly and easily. One of the most important features is the ability to develop disease-wide combination therapies; the more well-characterized samples we have to work with, the better combinations Cancer Math can develop. It just so happens we have dozens of characterized mouse models of sarcoma ready for Cancer Math modeling… more to come!
09/14/2017: Stress-testing project building code for the Cancer Math GUI. One of the key aspects of developing software is to make it accessible to everyone. Developing this front-end GUI to enable the whole lab to access Cancer Math algorithms quickly and easily. One of the most important features is the ability to develop disease-wide combination therapies; the more well-characterized samples we have to work with, the better combinations Cancer Math can develop. It just so happens we have dozens of characterized mouse models of sarcoma ready for Cancer Math modeling… more to come! 
09/27/2017: Last week was a busy week indeed! Aside from progressing on several scientific and programming projects, I also had the opportunity to visit our veterinary collaborators at the CSU Flint Animal Cancer Center in Fort Collins, Colorado. Dr. Bernard Seguin has been a wonderful collaborator over the past few years, and this was my first opportunity to present Cancer Math to the scientists and clinicians there. it’s exciting to lay out our vision for the future of personalized cancer treatment. You may be wondering what children’s cancer and animal cancer have in common… well, it turns out that dogs spontaneously develop the same sorts of tumors often seen in children, especially sarcomas. And since humans and dogs live side by side, we have long considered that dog cancer patients might be a great way to explore new treatments that can be translated to children’s cancer treatment. This presents a great way to test personalized cancer therapy built by cancer math… by testing it in dogs with cancer who also need new treatment options built for their biology. With a few successful tests in dogs, it will be that much easier for oncologists to explore using cancer math to help treat children’s cancer. See you soon, Super Sam foundation!
10/23/2017: Lots to report on since my last post! Biggest of all is my trip to the Super Sam Foundation Hope Gala. It was an unbelievable privilege to visit Matt, Cassie, Ava, and all the wonderful people who made the Hope Gala happen. It was great working alongside all of you on the last minute preparations to what proved to be a fun, fantastic, and heartfelt event. Sam’s story is powerful, and experiencing the emotion in the room firsthand is always a source of motivation for a researcher in the trenches. A big thanks to Ava for the lovely introduction, for all in attendance who listened to my presentation on my years of research, and for the support of everyone at the Hope Gala for supporting my work in changing the way children’s cancer is treated. (Full disclosure: this is probably the only picture I ended up with after the Hope Gala. I took quite a few Snapchats of the gorgeous venue for my friends, but I was a little too focused on my upcoming presentation to remember to keep any pictures for myself!).  On the science front, a lot of work has gone into wrapping up the manuscript that will be critical to the next stage of moving Cancer Math to clinical utility. Concomitantly, we have been planning strategies to bring Cancer Math into the clinic to meet the needs of those cancer patients seeking new options to continue their fight against cancer (and those patients with diseases lacking defined clinical treatments from the outset). Manuscript writing is always a time-consuming process, but alongside that has been working with our new scientist (Cora) on computational modeling of patient stratification for rhabdomyosarcoma, supporting the research on Osteopontin in rhabdomyosarcoma with Naren and Ali, and statistical analysis of in vivo experiments with Megan, and submitting two letters of intent for research projects ready for further development. I’m proud to work with and support my friends and colleagues. And coming up, we have some important experiments to determine if cell cultures we have developed are cancerous in behavior. More to come!
On the science front, a lot of work has gone into wrapping up the manuscript that will be critical to the next stage of moving Cancer Math to clinical utility. Concomitantly, we have been planning strategies to bring Cancer Math into the clinic to meet the needs of those cancer patients seeking new options to continue their fight against cancer (and those patients with diseases lacking defined clinical treatments from the outset). Manuscript writing is always a time-consuming process, but alongside that has been working with our new scientist (Cora) on computational modeling of patient stratification for rhabdomyosarcoma, supporting the research on Osteopontin in rhabdomyosarcoma with Naren and Ali, and statistical analysis of in vivo experiments with Megan, and submitting two letters of intent for research projects ready for further development. I’m proud to work with and support my friends and colleagues. And coming up, we have some important experiments to determine if cell cultures we have developed are cancerous in behavior. More to come!
11/06/2017: This week we welcomed a new potential collaborator into the fold, Dr. Stephen Ramsay at Oregon State University. Like me, he’s an electrical engineer working on cancer biology, and like cc-TDI, he’s working on sarcoma! In his case, it’s feline-origin cancer which makes a nice partner to our data on human and canine sarcoma. Seems there are some common biological signals amongst the three species, and it happens to be a biological pathway we’re currently exploring. There could be some interesting joint work going forward, and after a great couple of summer interns (Nick and Chandler) we’re excited by the coming possibilities of future collaborations with OSU. On the cancer math and cc-TDI front, the cancer math manuscript is going through final revisions before resubmission! This manuscript is critical to the continued development of Cancer Math. We’re almost almost there. The DIPG manuscript is also in the final stages as well. We’re counting on the Tech Transfer office at Hospital for Sick Children in Toronto to come through for us quickly. It was also a productive week on the grant front! Grants are one essential mechanism that helps support our research, and this past couple of weeks I submitted two letters of intent for promising research projects! It will be a busy grant cycle the next few months, so that means late nights in my favorite coffee shop. It’s ok, they know me (and Stormy!) there. Final thoughts, I’m starting a new project on the biology side. I’ll be transforming a mutant plasmid via E. coli to build up a large enough plasmid vector population to transfect into sarcoma cells and normal cells. Basically… giving sarcoma cells a rare mutation that will enable us to study how this rare mutation changes the behavior of the cancer. It’s a small but important project, and like most things in the lab, it’s motivated by a child struggling with a rare disease. picture credit: BioCat.com (https://www.biocat.com/cdna-clones/clones/RC403308-OR) 
11/22/2017:Thanksgiving is fast approaching, which always means everything in the scientific world slows down. Not really the case here at cc-TDI! This past week I’ve been focused on concluding the two key manuscripts and working on a few key Cancer Math grants, the most recent draft was sent for review today! Our associates at Beijing Genomics Institute, the organization to which we contract all of our physical sequencing work, provided us with a few critical data sets today, including those necessary for the final piece of the Cancer Math manuscript. I’m moving them to the servers where I will begin computational analysis via analysis pipelines on our high-performance computing servers. Results should be available tomorrow… which is indeed another workday for me (maybe half workday, I have a vegan Thanksgiving to attend). On the biology front, we’re starting xenograft (human tissue into mice) experiments to attempt to develop important cancer models for alveolar soft part sarcoma (ASPS) and Ewing’s Sarcoma. We should know in the next month or two whether the mice are able to grow tumors. Additionally, the RET in rhabdo project should be expanding in the near future, which will require educating myself in the particulars of lentivirus-based genome altering (using modified HIV virus to stably change the genome of human cells) in order to add a mutant gene into several models of human and mouse cancer cells. Finally, there are some exciting steps forward for supporting the continued development of Cancer Math. Nothing I can report on at present, but there should be some exciting news in the next few months!Have a great Thanksgiving holiday, everyone!
12/22/2017: The past few weeks have been a rush at cc-TDI. One of them a rush of a different sort, as I took a week’s vacation/journey to Scotland and Iceland. It was deeply needed after two years of no personal travel. Even in the middle of winter, the European North is a sight to behold. Please enjoy a couple pictures from my travels!  On the research side, the big announcement this week is resubmission of the Cancer Math manuscript! It’s been a long (but necessary) time coming. With the support of my friends and colleagues at cc-TDI, and all of my collaborators on the manuscript, the revised manuscript is in the hands of the editors and should soon be out for a review. If everything goes well, the manuscript will be published soon after the new year.
On the research side, the big announcement this week is resubmission of the Cancer Math manuscript! It’s been a long (but necessary) time coming. With the support of my friends and colleagues at cc-TDI, and all of my collaborators on the manuscript, the revised manuscript is in the hands of the editors and should soon be out for a review. If everything goes well, the manuscript will be published soon after the new year.  At the same time, our DIPG manuscript should be published in short order, which will lead into a great new research project on a promising new target (or target combination!) for the treatment of DIPG. We have received positive responses to letter of intent submissions, so we’ll be submitting to a couple grant opportunities coming up.
At the same time, our DIPG manuscript should be published in short order, which will lead into a great new research project on a promising new target (or target combination!) for the treatment of DIPG. We have received positive responses to letter of intent submissions, so we’ll be submitting to a couple grant opportunities coming up.  Meanwhile, several xenograft mice are growing to establish tumors for model development and experimentation. Everything is wrapping up as the year comes to a close, and everything is happily well timed. The new year will start fresh with several new projects and hopefully some further coding with Chandler and Nick, the summer interns! We are close to concluding our easy-to-use software pipeline. With my plate mostly clear, it’s time to delve deep into analysis and programming to push Cancer Math modeling forward, and resume my role as support to everyone at cc-TDI. Cancer Math’s next big role: helping Andy integrate newly generated sequencing data to identify promising drug combinations for classical and anaplastic Wilms’ Tumor! Happy holidays, everyone!
Meanwhile, several xenograft mice are growing to establish tumors for model development and experimentation. Everything is wrapping up as the year comes to a close, and everything is happily well timed. The new year will start fresh with several new projects and hopefully some further coding with Chandler and Nick, the summer interns! We are close to concluding our easy-to-use software pipeline. With my plate mostly clear, it’s time to delve deep into analysis and programming to push Cancer Math modeling forward, and resume my role as support to everyone at cc-TDI. Cancer Math’s next big role: helping Andy integrate newly generated sequencing data to identify promising drug combinations for classical and anaplastic Wilms’ Tumor! Happy holidays, everyone!
03/05/18: Last week at cc-TDI was filled with interesting visitors! Our first of the week was a molecular biology expert with years of experience working in pharma. His depth of experience in non-cellular biology is impressive and interesting to see. Our second visitor hails from Australia, and shared his work in machine learning approaches to challenges in clinical care. It’s fascinating work, and while I can’t share the details of unpublished work, it nicely serves a need before Cancer Math would come into play. I will keep an eye on upcoming publications, as I think there’s the potential for collaborations to bring computation to the clinic.
The cancer math manuscript will be resubmitted this week, just waiting on final overview from Charles!
Similarly, last week’s DIPG grant to St. Baldrick’s is mostly written and in local peer review. I’ll be submitting it shortly with plenty of time to spare. Everything should be coming to fruition soon.
Much of this past week was spent on working with sequencing data for my colleagues, finishing papers, finishing grants, and pushing forward on the GEAR project. Happily, the first round of GEAR experiments will begin this week, and Cora will soon be finalizing the eRMS dendrogram. Soon we’ll have some promising data-driven hypotheses to explore!
Thank you as always, SuperSam supporters, and please stay tuned!
03/23/2018: Hello SuperSam supporters!
This week I have the following updates on my progress on finding new treatments for high risk and metastatic RMS.
From the scientific side, I have been focused on identifying aRMS disease subtypes (known as endotypes) in conjunction with the GEAR research project. This project focuses on identifying distinct disease subtypes in aRMS to better target new therapeutic approaches. Similar to Cancer Math, we will move further away from the “one size fits all” approach still used clinically, and define new treatments based on the genetic and transcriptomic status of aRMS patients. We are currently exploring two aRMS endotypes: a RET-expressing aRMS subgroup and the PAX7 subgroup. The first set of experiments on the RET subgroup are detailed on the GEAR blog.
Additionally, I have been working closely with Naren on the data analysis side of the entinostat project to support the entinostat clinical trial for aRMS. In particular, we have been working to identify the mechanism of action necessary to understand why entinostat works to better target the drug to recurrent and refractory aRMS patients through our current clinical trial (ADVL1513).
From the publication side, we have reached two key milestones in the pursuit for deeper scientific and clinical understanding of aRMS. The Cancer Math manuscript for personalized therapy (validated in pediatric sarcomas, including aRMS) was resubmitted to a scientific journal for review, which brings us closer to the critical next steps in enabling Cancer Math to support clinical decision-making for deadly pediatric disease, especially metastatic aRMS.
And finally, I’ve been working with Dr. Stephen Ramsay at OSU to finalize a manuscript exploring the comparative oncology of human, canine, and feline sarcoma (through our collaboration with Dr. Bernard Seguin at CSU) which will help us better understand the biological commonalities in mammalian sarcoma, and to further understand the differences between non-RMS tumors and RMS tumors; understanding differences will help us better tailor treatments towards metastatic aRMS.
Thank you as always, SuperSam supporters, and please stay tuned!
03/28/2018:
Hello SuperSam supporters!
This week I have the following updates on my progress on finding new treatments for high risk and metastatic RMS.
From the scientific side, the GEAR project to find new treatments for aRMS subtypes is well underway. The first set of RET-positive aRMS drug validation experiments are now underway, with a second set beginning over the weekend. These experiments will help us understand the biology of this rare and dangerous subtype, and will also help us explore potential treatment options. We are driving for completion of the first round of single agent and combination agent validation studies in the next 2-3 weeks.
There are no major updates from my side on the critical entinostat work this week, though we will begin a major manuscript push shortly. I will play a critical role in the data analysis portion once the push begins.
From the publication side, the Cancer Math manuscript is under review, which is a promising step. The current home for the manuscript promises a fast turnaround, so we should hear back soon. In the meantime, I will be preparing another grant, this time for the Alex’s Lemonade Stand Foundation, focusing on validation of Cancer Math driven personalized therapy in multiple aRMS models.
Thank you as always, SuperSam supporters, and please stay tuned!
04/04/2018:
Hello SuperSam supporters!
This week I have the following updates on my progress on finding new treatments for high risk and metastatic RMS.
From the scientific side, the GEAR project to find new treatments for aRMS subtypes is barreling through the aberrant RET endotype subproject. Two of three replicates of both mouse cell line experiments have been completed. The final replicate will begin tomorrow, as will the first set of human cell line replicates. We should have a robust dataset in the next 1-2 week, as predicted. The next step on the available data is to begin formal analysis to confirm overall effect of single agents and combination agents. Completion of the first round of single agent and combination agent validation studies is on track. This dataset will be a cornerstone of the first GEAR manuscript
There are no major updates from my side on the critical entinostat work this week. Naren is finishing up a few key experiments on entinostat before we return to drafting new manuscripts.
From the publication side, the Cancer Math manuscript is still under review, hopefully we will be hearing back very soon. Having been under review for about 3 weeks, I would expect 1-2 more weeks before a decision. Writing of the next grant to support Cancer Math for metastatic and refractory aRMS is underway, and I aim to have the first major draft done in the next couple of weeks. It’s always a race, but the Cancer Math work is mature and ready for the next step.
Thank you as always, SuperSam supporters, and please stay tuned!
04/11/2018:
Hello SuperSam supporters! This week I have the following updates on my progress on finding new treatments for high risk and metastatic RMS.
From the scientific side, the GEAR project aberrant RET endotype subproject is more than halfway through the first data generation phase. All mouse replicates have been completed and analysis is underway, focusing on analyzing the effect of drugs working in combination. Human replicates are underway, though the first human experiment was not successful. Currently, two replicates of six will be completed this week, and two more should be started late this week or early this weekend. Formal analysis of the mouse data should be completed in the next two days. The mouse dataset, along with the human dataset, will guide the next steps in the GEAR project and the GEAR manuscript
The Cancer Math manuscript is still under review, hopefully we will be hearing back very soon. This morning, I presented the next major RMS-focused grant to my lab mates, who provided valuable feedback, which will strengthen the next grant applications for personalized RMS therapy.
Finally, I am also pleased to announce that we have officially published our research identifying a new promising target in DIPG, http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0193565 .
Thank you as always, SuperSam supporters, and please stay tuned! Noah
04/18/2018:
Hello SuperSam supporters!
This week I have the following updates on my progress on finding new treatments for high risk and metastatic RMS.
From the scientific side regarding alveolar rhabdomyosarcoma, the GEAR project aberrant RET endotype subproject is more than halfway through the first data generation phase. All mouse replicates have been completed and analysis is completed. Data visualization is also complete; part of my work this week was to generate simple tools to quickly and reliably produce clear and appealing visualizations of two-dimensional data. We will likely use that going forward for presenting data at cc-TDI. Human replicates are underway, with three of the six having gone through data generation. Due to concerns about robustness, I am analyzing the recently generated data to determine if it meets our standards. Any datasets which do not will result in additional replicates, which is part and parcel of working in molecular biology. At cc-TDI, we accept nothing but the most robust data when defining new treatments for children’s cancer. Formal analysis of the mouse data should be completed in the next two days. The mouse dataset, along with the human dataset, will guide the next steps in the GEAR project and the GEAR manuscript
Additionally, studies of the PAX7:FOXO1 ARMS variant is spinning up as one of the endotypes in ARMS. The PAX7 variant is not generally as deadly as the PAX3 variant, but its rarity and lack of models makes it difficult to study. We will accept the challenge nonetheless.
The Cancer Math manuscript is still under review, hopefully we will be hearing back very soon.
Thank you as always, SuperSam supporters, and please stay tuned!
04/25/2018:
Hello SuperSam supporters!
This week I have the following updates on my progress on finding new treatments for high risk and metastatic Rhabdomyosarcoma.
From the scientific side, the GEAR project aberrant RET endotype subproject the first data generation phase is nearing completion. All mouse replicates and analyses, and the experimental replicates for one of the two human cell lines is almost complete. Many screening plates for the human experiments were re-printed to ensure experimental robustness. One cell line had higher than expected growth rates in the petri dish, and I am concerned the high cell population may have affected the results. I am running repeat experiments to confirm whether or not this is the case, though this will not delay results much. The mouse dataset, along with the human dataset, will guide the next steps in the GEAR project and the GEAR manuscript
Additionally, studies of the PAX7:FOXO1 ARMS variant is spinning up as one of the endotypes in ARMS. We received a cell line for this very rare cancer from overseas collaborators, and unfortunately the culture (which was sent live) does not seem to be recovering well. I will follow up next week to have a cryopreserved sample sent to us.
The Cancer Math manuscript is reviews came back and we are now making the few requested changes. Hopefully publication soon!
Thank you as always, SuperSam supporters, and please stay tuned!
05/02/2018:
Hello SuperSam supporters!
This week I have the following updates on my progress on finding new treatments for high risk and metastatic RMS.
From the scientific side, the GEAR project aberrant RET endotype subproject the first data generation phase is nearing completion. All mouse replicates and analyses, and the experimental replicates for both human cell lines are almost complete. The previous concerns about cell population and the effect on experimental outcome have been resolved, and the final replicates are completing now. Analysis of the data will begin shortly and should conclude next week. The mouse dataset, along with the human dataset, will guide the next steps in the GEAR project and the GEAR manuscript
The PAX7:FOXO1 ARMS cell line from our overseas collaborators did not recover, and the collaborators should be sending us cryopreserved samples sometime this week or next week.
The Cancer Math manuscript’s reviews came back and the requested changes are almost complete. We will be resubmitting soon.
Thank you as always, SuperSam supporters, and please stay tuned!
05/10/2018
Hello SuperSam supporters! This week I have the following updates on my progress on finding new treatments for high risk and metastatic RMS.
From the scientific side, the GEAR project aberrant RET endotype subproject the first data generation phase is nearing completion. All human replicates are now complete, and the data analysis is under way. Analysis should be complete in the next couple of days. The mouse dataset, along with the human dataset, will guide the next steps in the GEAR project and the GEAR manuscript
The PAX7:FOXO1 ARMS cell line from our overseas collaborators did not recover, and the collaborators are sending us cryopreserved in the next couple of weeks. Having cryopreserved cells should make recovering cells significantly easier.
The Cancer Math manuscript is reviews came back and the requested changes are almost complete. We will be resubmitting soon.
05/17/2018
This week I have the following updates on my progress on finding new treatments for high risk and metastatic RMS.
From the scientific side, the first stage in the GEAR rhabdomyosarcoma project is complete. All human and mouse data has been analyzed, and we are now pursuing a deeper understanding of the role of RET mutations and aberrant expression in aRMS. We will be establishing new stable cell line models this coming week.
The PAX7:FOXO1 ARMS cell line from our overseas collaborators did not recover, and I am currently setting up a shipping contract with World Courier to have cells on dry ice sent to us.
The Cancer Math manuscript is reviews came back and the requested changes are complete. I am now sending the manuscript through a final review with my lab mates, and it will be resubmitted vert soon! Hopefully in the next few days.
Thank you as always, SuperSam supporters, and please stay tuned!
update 6/06/2018:
By popular demand, we are switching to less-frequent, more content-rich monthly posts. These will appear as new blog posts on our website. For exclusive! week to week updates of late-breaking results, email molly@cc-tdi.org to be subscribed. Thank you for supporting this rhabdomyosarcoma research!

